Genetic Genealogy of the Milam / Mileham / Milum Surname
The Origin of Surnames in Great Britain
Before the Norman Conquest of Britain, people did not have hereditary surnames: they were known just by a personal name or nickname. After 1066 AD, the Norman barons introduced surnames into England and the practice gradually spread. Initially, the identifying names were changed or dropped at will but eventually they began to stick and to get passed on, so trades, nicknames, places of origin, and fathers' names became fixed surnames. By 1400 most English families and those of Lowland Scotland had adopted the use of hereditary surnames.
Surnames deriving from a place are probably the oldest and most common. They can be derived from numerous sources - county, town or estate - or from features in the landscape - hill, wood, meadow, brook, lake. Many of these names and their derivation are obvious, others less so. The names Pickering, Bedford, Berkley and Hampshire might have been the names of estates on which the individuals worked and lived. If a person migrated elsewhere, their former place name would be a convenient way to distinguish them from others with the same first name.
The oldest known record of a MILAM in England is from the County of Norfolk: the 1375 AD Will of George de Mileham. The translation of de from French is of in English. So George de Mileham literally meant George of Mileham - which indeed was, and is, a village in Norfolk. William the Conqueror granted the Mileham lordship - a name of Anglo-Saxon origin - to Alan Fitz-Flaald (son of Flaald), ancestor to the Earls of Arundel. [614, 615]
The origin of surnames may explain why Englishmen with a MILAM surname are of different genetic ancestories i.e. not one single genetic family but rather different genetic families perhaps once living in, or around, the village of Mileham who adopted its name. Some of them were of Germanic (Angles, Jutes, Saxon) origin, others of Scandinavian (Viking / Norman) origin and perhaps others of Celtic or even Roman origin. [623, 631] Each of those ethnic groups represent foreign invasions which displaced the early Britons to the west and north since the invasions occured by sea mostly along the east and southeast coasts. [616, 635] In 2021 between the United Kingdom and the United States, we know of eight genetically distinct MILAM families.
Basic Concepts in Genetic Genealogy
Like all surnames, the MILAM surname was passed on from father to son, father to son over centuries like the male Y chromosome. The human genome (link) has 23 pairs of chromosomes (link), one half contributed by the mother's egg (ova) and one half contributed by the father's sperm. Since the mother has two X chromosomes, she always contributes an X. Since the father has an X and a Y chromosome, he may contribute either which determines a child's gender. Since only males have the Y-chromosome, only men and their Y-DNA test results can be utilized in genealogical surname research. [617]
To make things more complicated, during the development of a sperm or an ova each member of a chromosome pair swaps genetic segments (re-combines) with the other including to a limited extent the X and Y chromosomes. Fortunately, there is a large portion of the Y chromosome which never recombines - the non-recombining portion - and this is the part which is analyzed in genetic Y-DNA testing. The chromosomes from the nucleus of a dividing human cell look like this:
 |
Y-DNA Testing for Genealogy
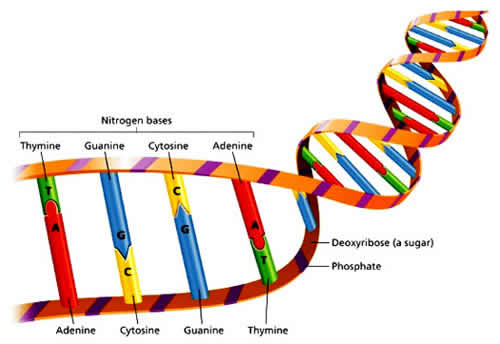
Each chromosome is made up of DNA strands tightly coiled many times around proteins called histones that support its structure. Nucleotides are nitrogen bases which make up the so-called "alphabet" of DNA. But there are only four "letters" in the DNA alphabet. These nitrogen bases or nucleotides are adenine (A), thymine (T), guanine (G) and cytosine (C). A always pairs with T, while G always pairs with C. These linkages are termed base pairs.
 |
Although a large portion of the Y chromosome is non-recombining and is transmited virtually unchanged from father to son occasionally mistakes in the copying process occur which are called Single Nucleotide Polymorphism (SNP pronounced snip) or variants. These are not technically mutations since they donot result in any deficiency or change in function. More than 55,000 SNP's have been identified which can differentiate the various paternal lineages in the world.
Each SNP that occurs in a new individual is inherited by all his descendants. This is why men descending from the same male ancestor share the same series of SNPs, or variants, inherited from all of their accumulated paternal ancestors for thousands of years (patrilineal inheritance). Men with the same set of inherited variants can therefore be classified in the same genetic family which population geneticists call a Haplogroup (link) [630]. By testing for these variants, it is possible to trace one's genetic genealogy.
Short Tandem Repeat (STR) Tests
All chromosomes contain sequences of repeating nucleotides known as Short Tandem Repeats (STR). The STR segments occur in what is considered to be non-coding DNA or "junk" DNA - an area with no instructions for producing anything. STR regions on the Y chromosome are designated by a DYS number and are often referred to as a genetic marker. For example, DYS522 has repeats of the nucleotide sequence G-A-T-A which may be repeated 8 to 17 times. The number of repetitions at a specific DYS region varies from one person to another; and the number of repetitions provides the value for that DYS marker. Here is what DYS522 results might look like for four different men:
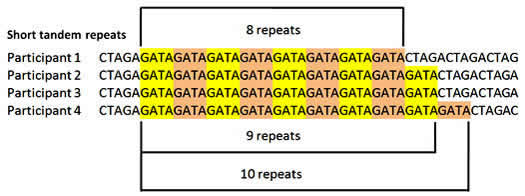 |
Genealogical STR (link) testing involves analyzing the STR segments on the Y chromosome which are referred to as Y-DNA tests. Family Tree DNA offers Y-DNA tests for 37, 67 and 111 markers named YDNA-37, YDNA-67 and YDNA-111.
Below are the results for 4 men who took a hypothetical YDNA-19 test. The first marker DYS393 repeats the nucleotides A-G-A-T. All persons in this example have a value of 13 for DYS393. If we look at the second marker DYS390, one person has a value of 23 and the others have a value of 24. In this table of results, values below the mode (the most frequent result) are in a blue box; values above the mode are in a pink box. The value 23 for DYS390 is in a blue box since the mode was 24. Similarly for DYS449 at the far right, the mode was 28 and the result of 27 is in a blue box. For DYS439 the mode was 12 therefore the values of 11 are in blue boxs.
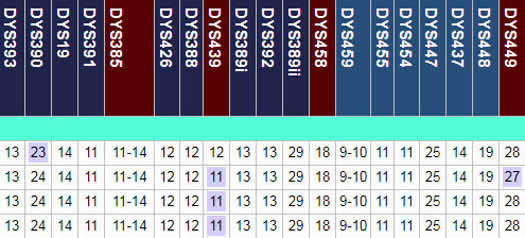 |
STRs results can provide a prediction of an individual's haplogroup although his haplogroup can only be confirmed by testing for that haplogroup's specific inherited variants, SNPs, acquired over millennia by his patrilineal line.
STR results also predict how closely two men are related. For example, if the results of a 37 marker YDNA-37 test are identical (genetic distance of 0), then there is a 50% probability that the two men's common ancestor lived no longer than two generations ago and a 90% probability that the ancestor lived no longer than five generations ago. [629] If their results differ by one (genetic distance of 1), then there is a 50% probability that the two men's common ancestor lived no longer than four generations ago as shown in this table.
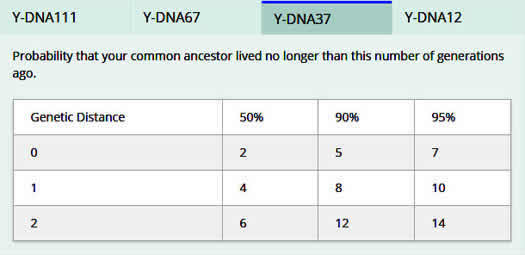 |
STR tests are the oldest type of DNA test and have been used for paternity, forensic and criminal investigations for several decades.
Single Nucleotide Polymorphism (SNP) Tests
A single nucleotide polymorphism (SNP) is a variant in a single nucleotide base pair of a DNA strand as illustrated below. It's basically a copying error which occurs during
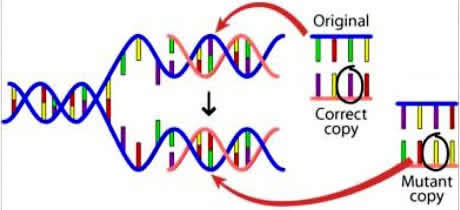 |
spermatogenesis whereby one nucleotide is substitued for another. Technically these are not considered mutations since there is no change in function associated with a SNP. SNP is pronounced "snip". More information on SNPs here (link) .
SNP variations are very stable which makes them ideal for marking the history of the male, or patrilineal, genetic tree. SNPs are named with a letter code and a number. The letters indicate the laboratory that discovered the SNP and the number indicates the order in which it was discovered. For example, BY100 is the 100th SNP documented by Big Y testing at Family Tree DNA which uses the code letters BY for its lab. Many SNPs have been named by more than one testing laboratory so often a SNP is represented by two alphanumeric symbols side by side.
A terminal SNP is simply the latest SNP of a male line which defines a new branch (subclade) which is known from current SNP testing. ("Terminal" as in bus terminal where lines branch; not terminal as in final branch!) A Haplotree is a growing thing because of more men BIG Y testing. You may have one terminal SNP today but when your branch of the tree grows due to more men testing you may have a different terminal SNP. This is why FTDNA, Williamson's The Big Tree and Y-Full's YTree update their trees every couple of months.
As mentioned above, STR test results provide a predicted haplogroup. But a panel of SNP test results are required to confirm a haplogroup. SNP testing also enables researchers to learn more about our deep ancestry going back thousands of years. The success of ancient DNA SNP testing of thousands of skeletons from ancient, pre-historic cultures has revolutionized our understanding of the movement of ancient peoples into Europe and, for that matter, into North and South America as well as into Indonesia and Australia.
Genetic Genealogy of the Virginia Milam
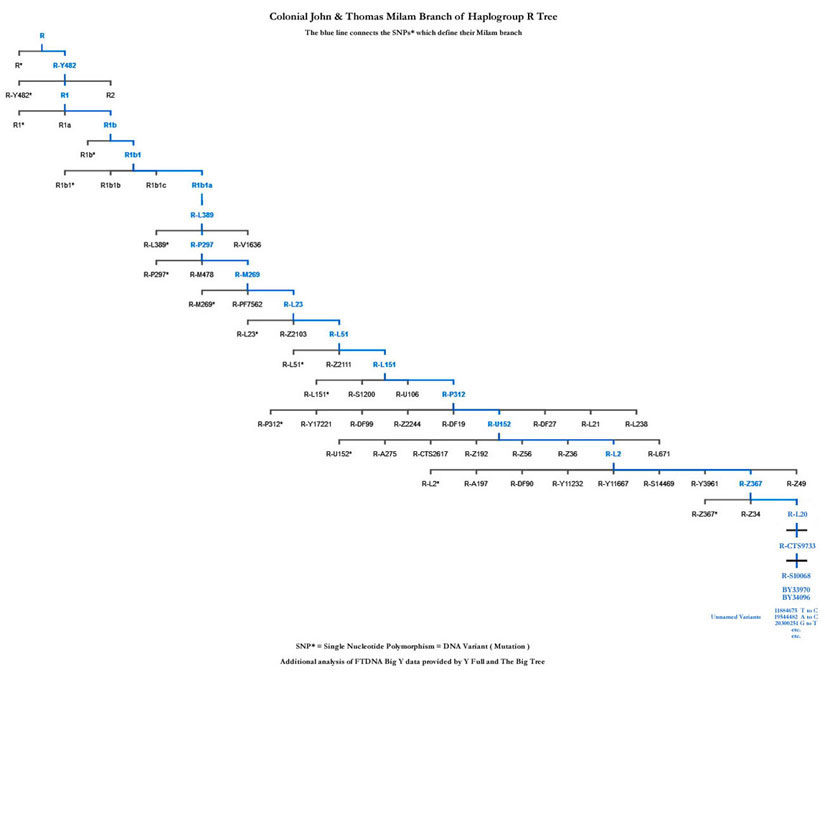
Fifty-three of the fifty-five American Milams tested belong to the same haplogroup (link) and are all descendants of John Sr and Thomas Milam who lived in the Piedmont region of the Colony of Virginia in the mid 1700s. On short tandem repeat (STR) testing their results are always predicted haplogroup R-M269. Haplogroup R (link) is one of the 20 major haplogroups of mankind. Its branch R1b accounts for ~ 95% of present day west European men. R-M269 is a dependable ancient SNP which defines most living R1b men; it first occured more than 13,000 years ago.
On the first panel of single nucleotide polymorphism (SNP) tests ( the M269 "Backbone" SNP Pack of 137 SNP markers ), the result for these Milams is SNP Z367. When one tests the advanced SNP panel ( the Z367 SNP Pack of 122 additional markers ), their terminal SNP is CTS9733. Imagine an inverted tree with branches descending downward: each SNP panel tests markers further down the branching tree. When one tests down to the furthest known branch of the tree, it is referred to as the terminal SNP of a person's patrilineal line.
The John Sr and Thomas Milam branch can be better visualized on the tree below. Please click on this chart to see a larger version. The blue line connects the major SNP mutations which were accumulated by their Milam paternal ancestors over thousands of years which define their Milam line.
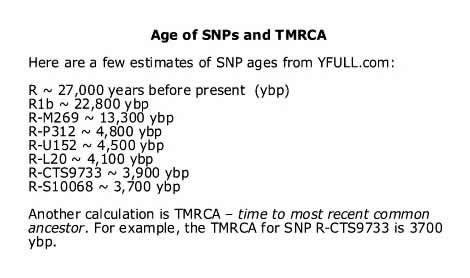
Yes, their SNP mutations did accumulate over many thousands of years. Here is a list of some of their major SNPs and the estimated time the variant first occured [618] :
 |
The Family Tree DNA "BIG Y" Chromosome Sequencing Analysis
When our first two BIG Y results were reported nearly five years ago, our Terminal SNP was CTS9733 and we each had 35 Unnamed Variants. Thanks to the thousands of men who have tested since and our Project's testees, most of those Unnamed Variants have been given official SNP names.
By April 2021 fifteen Americans from our Milam / Mileham / Milum Surname Project have taken the FTDNA BIG Y sequencing analysis. This analysis looks at more than ten million nucleotide base pairs on the Y-chromosome for SNPs or variants (see SNP discussion above). These fifteen descendants share more than 2000 SNPs in common representing the accumulated variants on their Y chromosome passed down to them by all their paternal ancestors over thousands of years.
All descendants of Thomas Milam are positive for these major SNPs:
Somewhere along his line, John Milam Sr acquired two additional SNPs which Thomas and his descendants do not have: BY67643 and BY184340. All of John's descendants therefor carry these major SNPs:
Were John Milam Sr and Thomas Milam Brothers?
In 2020 our Surname Project undertook to answer this question by recruiting eight additional John Sr and Thomas Milam descendants to order the FTDNA BIG Y 700 DNA sequencing analysis. It is not unusual that one new SNP might occur in one brother at his conception but not in the others. Indeed, if you look at the left side of my chart below, that is what happened with Thomas’s son, Rush, who acquired the new SNP BY34096 which his brothers did not acquire. By the way, his new SNP BY34096 defines a new genetic branch (subclade) in the Thomas Milam line and is referred to as a Terminal SNP.
What would be very unusual is for one brother to acquire two new SNPs at his conception. This is because a random Single Nucleotide Polymorphism (SNP) on the Y chromosome occurs about every 83 years on average, or once every 3 generations. Therefore, it would be quite rare for two new SNPs to occur during one conception. The finding that John Sr has two SNPs which Thomas hasn't strongly suggests that John and Thomas were not brothers. Proof of this came when www.yfull.com (link) completed their analysis of our BIG Y testees' results. Y-Full estimated that the Time to the Most Recent Common Ancestor (TMRCA) for John Sr and Thomas Milam was about 400 years before present (ybp) or around 1600 AD. That is when their Y-DNA began to diverge because of new SNPs or variants in the John Sr's line. This estimate is approximate but confirms that they were not brothers.
Historical facts also suggest they were not brothers: Thomas is first found in 1738 in Orange County and John Sr first appears in 1750 in Brunswick County, some 160 miles distant. These facts suggest that they may have arrived in Virginia at different times and settled in different places rather than arriving together as a family unit.
My chart below illustrates the genetic relationships between Thomas Milam and John Milam Sr and their sons. I use blocks containing the individual’s name and their inherited terminal SNP. The lines of descent are shown by arrows and I indicate when a new Terminal SNP was created. ("Terminal" as in bus terminal where lines branch; not terminal as in final branch!) Please click on this chart to see a larger version.
The chart shows that Thomas Milam inherited the same terminal SNP BY33970 as their British ancestors - no new SNP was created at Thomas’ conception. Thomas’ sons Benjamin, John and Zachariah inherited his Terminal SNP BY33970 - no new SNP for them either. However, Thomas’ son, Rush, acquired a new terminal SNP BY34096 at the time of his conception.
www.yfull.com (link) estimated the TMRCA for Rush Milam's Terminal SNP R-BY34096 was about 200 ybp - around 1800 AD - again a rough estimate.
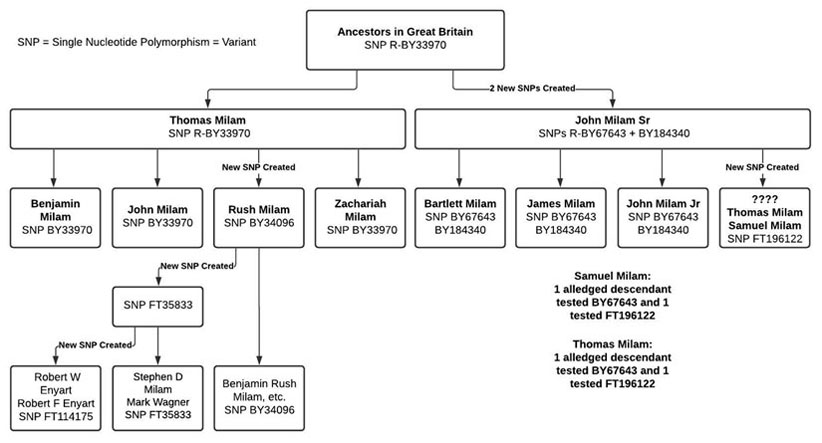 |
The left side of the chart also illustrates that the two Enyart / Milam men are descendants of Rush Milam via an unknown descendant who was the first Milam to acquire the terminal SNP FT35833. Zack Daugherty, our Surname Project co-administrator, suspects this man may be his “brickwall” Milam, possibly Rush Milam’s son, John. Later the Enyart / Milam line acquired another terminal SNP FT114175.
The chart also shows that the John Milam Sr sons Bartlett, James and John Jr inherited his two SNPs: BY67643 and FT184340. However, the two alleged descendants of Samuel Milam have different results: one alleged descendant tested BY 67643 and FT184340; the other tested the a new SNP FT196122. The same is the case for the two alleged descendants of Thomas Milam. So we don't know which of John Sr's sons acquired the new SNP FT196122 at his conception.
These different Y-DNA results are NOT a Y-DNA testing error. Rather the error is in the descendants' genealogical "paper trails". If we had two or more testees for other sons of Thomas or John Sr, we would probably see other inconsistencies in results due to errors in paper trails because it is very difficult to be certain of our ancestors from 6 or 7 generations ago.
Clarification
Apparently this chart confused some because it shows Benjamin Rush Milam as a descendant of Thomas' son, Rush. Some thought that I was referring to Moses' son, "Ben Milam" of Alamo fame. But Thomas' son, Rush Milam, also named a son, Benjamin Rush Milam, most probably to honor his brother, Benjamin, who died in our Revolutionary War. And there should be no doubt that the name "Rush" pays respect to his mother, Mary Rush, the daughter of William Rush whose farm was less than a mile from Thomas Milam's farm in Culpeper County. Details here (link).
Please remember that Thomas Milam's sons would have known their Rush grandparents, aunts and uncles up till 1761 when they moved to Bedford County. Really they were the only family his sons knew since Thomas' family was in England.
Acknowledgement
I wish to acknowledge Dr Zack Daugherty’s invaluable help in analyzing our FTDNA BIG Y results. Because of his maternal Milam ancestry, Zach is interested in our project and has recruited two new project members to Big Y tested with us. I asked Zack to join our project as an administrator two years ago because of his background in genetics.
Other Milam / Mileham / Milum families are on other branches of haplogroup R and also on branches of the haplogroups E, I and J. In 2021 between the United Kingdom and the United States, we know of eight genetically distinct MILAM families. I will now discuss what we learned from the Y DNA testing of 26 British men.
The Genetic Genealogy of the British Mileham / Milam / Milum
In 2006 - 2007 Lonzo Oliver Milam of Raleigh, North Carolina, and Cyril Alfred Mylam of London, England, recruited nine British men to join the Family Tree DNA Milam Surname Project and tested their YDNA. During 2017 we were fortunate to recruit an additional 17 British men for a total of 26. We focused our research on counties where we knew MILAMs were born in the 16th and 17th centuries as illustrated on these maps:
The men were initially tested with the YDNA-37 Short Tandem Repeat (STR) test which showed that most men fell into two large haplogroups: I1-M253 and R1b-M269. As the number of results grew, it was appropriate to selectively test some members further with Single Nucleotide Polymorphism (SNP) panels of 120+ SNPs. A few of these men were tested with a second more advanced SNP panel.
In 2019 we know that there are at least six genetically distinct MILAM families in Great Britain as may be seen by their YDNA results at our FTDNA webpage, here (link) .
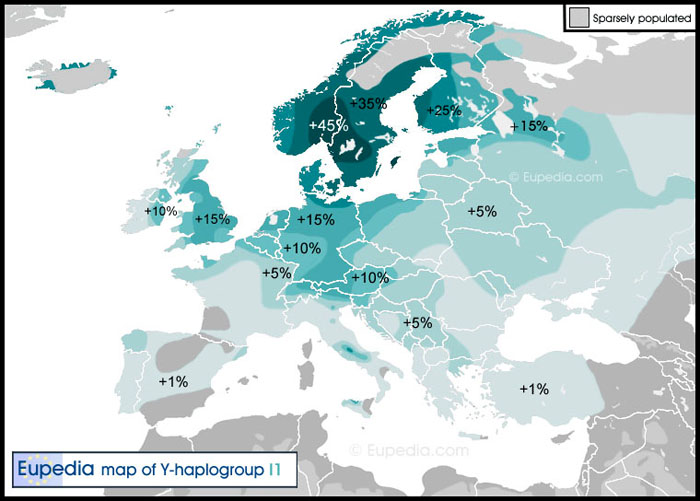
More than half of them (15) belong to our Great Britain-1 subgroup, haplogroup I1-M253>F2642, which among present day Europeans has a “Nordic” distribution. [619] Their oldest known ancestors were from eight different counties stretching from Lanarkshire in Scotland to Devon and Kent in the south. But they are concentrated in and around Berkshire (3), Hampshire (4) and Surrey (3). These are the men who are the same haplogroup as American Kit # 120119: I-M253>F2642 . This American's oldest known ancestor was William Mileham born ~ 1756 in the Colony of Pennsylvania.
 |
Five men belong to our Great Britain-2 subgroup, haplogroup R1b-U106>Z326, which among present day Europeans has a distinctly “Germanic” distribution. [620] Their oldest known ancestors were from Norfolk, Suffolk and Cambridgeshire not far from the village of Mileham with its Anglo-Saxon name and Norman ruins - a metaphor for the MILAM mix of ancestry.
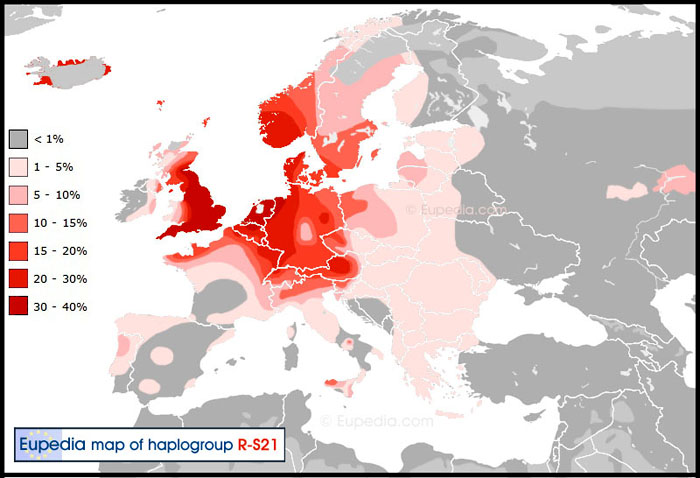
Our Great Britain-3 and Great Britain-4 subgroups belong to other branches of the R1b>U106 (S21) tree i.e. R1b-U106>L48 and R1b-U106>Z18 respectively and share the same “Germanic” distribution as seen on the map below. SNP U106 is also known as S21.
 |
Our Great Britain-5 member is of haplogroup R1b-L21 which is referred to as “Atlantic Celtic” and represents the peoples who brought the Copper / Bronze Ages to the Isles from the Rhine River Valley around ~ 2450 BC. [622, 630] They overcame the megalithic “Stonehenge” hunter-gathers and early farmers and largely replaced them becoming widespread throughout the Isles by the time of the Roman conquest. See the map below. [620, 621, 631, 632 ]
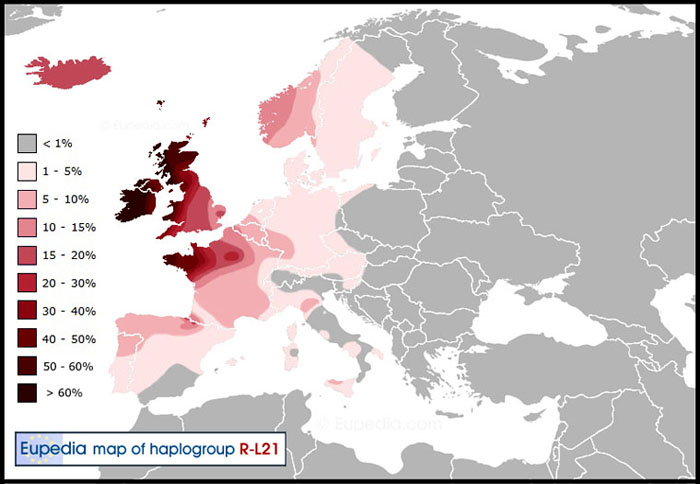 |
By comparing the three haplogroup distribution maps above, you can visualize that both in Great Britain and on the Continent the R1b-L21 peoples were displaced westward by Germanic and Nordic invaders after the fall of the Roman Empire. [623] Today haplogroup R1b-L21 makes up the majority of men in Ireland, in western Wales and Scotland, and in Brittany, France - all Celtic speaking areas. [616, 630, 634] DNA studies of hundreds of ancient skeletons have confirmed these population movements as discussed in these recent BBC Science articles:
Our Great Britain-6 member is of haplogroup J-M172 (also called J2) which has a distinctly Middle Eastern and Mediterranean population distribution, representing up to 30% of males in parts of Italy as shown below. One of the five Roman soldiers excavated at the York, England, archaeological site was haplogroup J2 (200 - 300 AD). [624, 625, 628, 635]
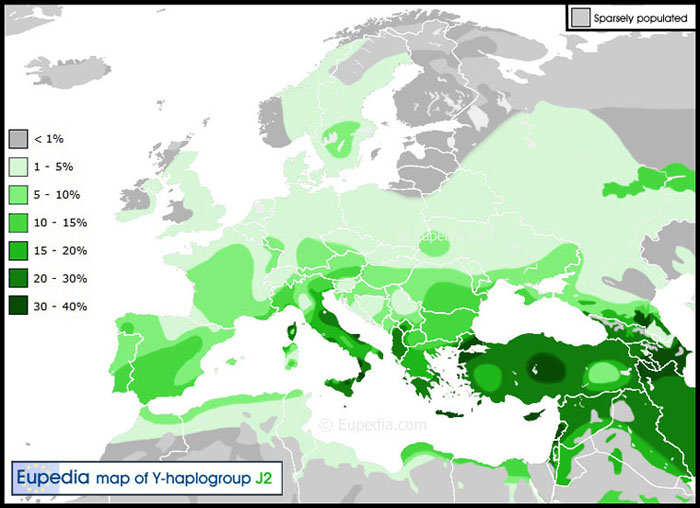 |
Alas, none of the 26 British men we tested has the same haplogroup as we Virginia MILAMs, R1b-U152 (S28). Ancient Y-DNA analysis on one of the five Roman-Britons excavated at Roman York ( Eboracum ), England, (DRIF-22) found him to be in our haplogroup R1b-U152-L2. [628] { More recent and deeper analysis of DRIF-22 found him to be: U152>L2>FGC22501>Y37744>A12416>BY3497. } He was one of the decapitated Roman era skeletons thought to be gladiators (slaves). Archeologists described him as 28-45 years and found he had a penetrating injury of his decapitated occipital skull. Carbon isotopes dated him to 100 to 400 AD. They concluded he was from a continental Celtic community probably in France or Belgium.
Researchers think that many R1b-U152 persons arrived in England with the Iron Age Hallstatt Celts (650 - 500 BC ) but thus far no ancient DNA has been tested for them. [626, 627, 633]
R1b-U152's highest representation in present day England (10 - 15%) is in the Norfolk / Suffolk region, around London and in the southeastern County of Kent as shown on the map below. Curiously across the English Channel is the village of Millam in the Nord-Pas de Calais region of France south of Dunkirk. Our haplogroup R1b-U152 is fairly high (15 to 20% of the population) in Northern France and Flanders as it is in County Kent. Unfortunately we were able to test only one Englishman whose ancestor was from Kent.
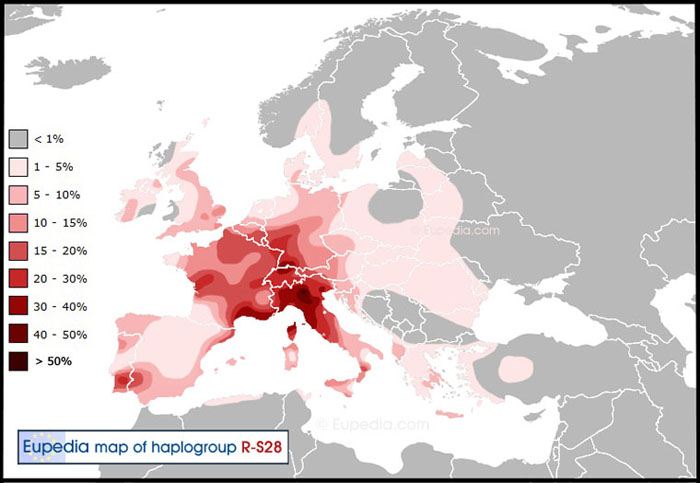 |
SPECULATION: After these negative results for finding a Y-DNA genetic match, we must consider the possibility that John and Thomas Milam of colonial Virginia were orphans taken in by a Milam family or perhaps their mother's family. It was not uncommon in the early 18th century when the English economy was in a depression and jobs were difficult to find that orphans viewed life in America as an indentured servant (link) to be their best option. This might also explain why the earliest records for John Sr in Brunswick / Chesterfield Counties and Thomas in Orange / Culpeper Counties place them ~ 165 miles apart: their indentures (contracts) were sold to different "masters" when they arrived in Virginia - or they arrived at different times. My best guess for the origin of John and Thomas Milam is near the bottom of my article on Milam Mariners and Merchants from Great Britain which you may read here (link) .
You may read the evidence for why they were indentured servants here (link) . And you may read my speculation that they embarked for Virginia from the Port of Whitehaven, England here (link) .
Here are links to articles with much more information about the haplogroups I1, R1b and J2:
This is the link to a discussion of the ancient DNA studies of the Roman soldiers found at the cemetery in Roman York ( Eboracum ), England:
William F. Milam, MD